Biacore X100 reference portal
Procedure for immobilization pH scouting
While the general recommendation to immobilize proteins at pH 4.5 works adequately in many cases, the coupling conditions frequently need to be optimized for best results. In order to determine suitable coupling conditions without permanently modifying the sensor chip surface, inject a short pulse (e.g. 2-3 minutes) of ligand in coupling buffer over a surface that has not been activated with EDC/NHS. Pre-concentration of the ligand on the surface will be seen as an increase in response, and will give an indication of whether the conditions are suitable.
A procedure for determining a suitable coupling pH is outlined below. An analogous procedure may be used to test other aspects of the coupling conditions.
- Prepare ligand solutions in the different coupling buffers to be tested. As a general rule, cover the pH range 4–5.5 in steps of 0.5 pH units for initial scouting.
- Inject the ligand solution (2-3 minutes). Start at the highest pH to reduce the risk of aggregation or precipitation of the ligand on the surface.
- Set report points just before the start and end of the injection to determine the level of electrostatically bound ligand.
- Inject a short pulse (e.g. 1 minute) of 1 M ethanolamine-HCl pH 8.5 (included in the Amine Coupling Kit) or 50 mM NaOH over the surface to remove the last traces of ligand.
- Repeat steps 2–4 with ligand in different buffers.
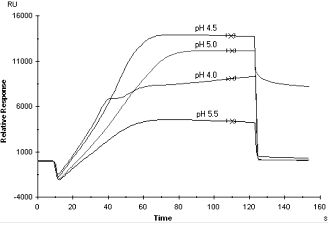
An example of pre-concentration scouting results. Binding increases as the pH is reduced from 5.5 to 4.5. At pH 4.0, the sensorgram is irregular and bound material does not dissociate from the surface at the end of the injection, indicating that the protein is aggregating or denaturing. The optimum pH for this protein is 4.5.
In assessing the results of pre-concentration tests, bear in mind that pre-concentration is generally more efficient at lower pH values, but the amine coupling chemistry requires uncharged amine groups and is therefore more efficient at higher pH.
Many protein ligands tend to aggregate or precipitate at low pH. This is often visible as irregularities in the sensorgram: avoid using buffers in which this behavior is observed. If you do not obtain satisfactory electrostatic pre-concentration at pH 4.0-5.5, try increasing the pH value. Try increasing the ligand concentration and if possible reduce the ionic strength of the buffer if the ionic strength is high in the ligand stock solution. If this does not help, you may need to use a different coupling approach.
Lab protocol
Lab procedure Preparation of Biacore immobilization buffers of different pH is available for download on www.cytivalifesciences.com/solutions/protein-research/Interaction-analysis-with-Biacore-surface-plasmon-resonance-SPR/Get-started-with-surface-plasmon-resonance-SPR-interaction-analysis
Immobilization of macromolecular ligands is usually performed from reasonably dilute ligand solutions (10–50 µg/ml or less), and result in immobilization levels that correspond to a concentration in the dextran matrix of the order of 10–20 mg/ml or more 1. This is achieved through electrostatic pre-concentration of ligands in the dextran matrix.
At pH values above about 3.5, the carboxymethylated dextran on the sensor chip surface is negatively charged, and electrostatic attraction provides an efficient means for concentrating positively charged ligands on the surface. This mechanism works well for most proteins and for several other kinds of biomolecules.
The primary requirement for this electrostatic pre-concentration on the surface is that the pH of the ligand solution should lie between 3.5 and the isoelectric point of the ligand, so that the surface and the ligand carry opposite net charges. In addition, the electrostatic interactions involved in pre-concentration are favored by low ionic strength in the coupling buffer. In general, covalent immobilization of proteins is typically performed from solutions in 10 mM buffer (e.g. sodium acetate) at pH 4–5.5. A range of ready-to-use buffers for protein immobilization is available from Biacore. The extent of pre-concentration of ligand on an unactivated sensor chip under different buffer conditions can be measured to determine the suitable conditions of pH for immobilization.
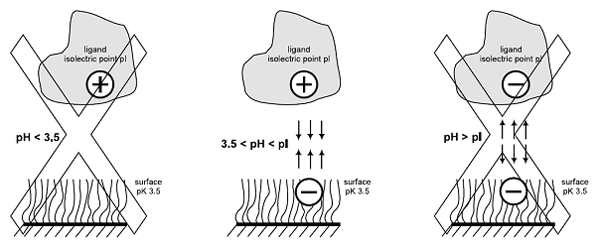
Ligand is concentrated on the surface through electrostatic attraction when the pH lies between the isoelectric point of the ligand and the pKa of the surface. If the pH is too low or too high, ligand will not be concentrated on the surface.
Some ligands (for example highly acidic proteins and nucleic acids) are negatively charged even at low pH values and cannot be efficiently pre-concentrated on carboxymethyl dextran surfaces. Modification of the ligand can in some cases help to reduce the acidity: for example introduction of active disulfides for thiol coupling may raise the isoelectric point of proteins sufficiently to allow pre-concentration. High affinity capture is an alternative immobilization approach that does not depend on electrostatic pre-concentration: attachment of biotinylated nucleic acids to streptavidin-coated surfaces is one example. Capturing interactions can be exploited in buffers of physiological ionic strength or higher to reduce the effects of electrostatic repulsion of acidic ligands.
1 An SPR response of 1,000 RU corresponds approximately to a surface concentration of 1 ng/mm2 for an average protein ligand on Sensor Chip CM5. If the thickness of the dextran matrix is taken to be 100 nm, this is equivalent to a volume concentration in the dextran matrix of 10 mg/ml.
For many proteins, coupling in 10 mM acetate buffer pH 5 works well. If you need to use other conditions, bear the following considerations in mind:
- The buffer pH should be at least 0.5–1 unit below the isoelectric point of the ligand. For ligands with isoelectric point above pH 7, the buffer pH may be increased to 5.5 or 6. The optimum pH can be determined experimentally.
- The ionic strength should be low (recommended 10–20 mM monovalent cations).
- Buffer components containing primary amine groups and other strong nucleophilic groups (e.g. Tris, sodium azide) must be avoided for amine coupling, since these will compete with the ligand for activated esters on the sensor chip surface. Thiol coupling must be performed in the absence of reducing agents.
See also
Panel 2 header
Lorem ipsum dolor sit amet, consectetur adipisicing elit. Repellendus aliquid tempore natus voluptates repudiandae, eos dolorem libero inventore, quod incidunt, asperiores. Reiciendis itaque enim pariatur, blanditiis minima autem quo a nulla, obcaecati quis, excepturi atque ab rerum! Sequi molestias vel eum, hic, perspiciatis eius suscipit reprehenderit molestiae vitae similique.
Panel 3 header
Lorem ipsum dolor sit amet, consectetur adipisicing elit. Repellendus aliquid tempore natus voluptates repudiandae, eos dolorem libero inventore, quod incidunt, asperiores. Reiciendis itaque enim pariatur, blanditiis minima autem quo a nulla, obcaecati quis, excepturi atque ab rerum! Sequi molestias vel eum, hic, perspiciatis eius suscipit reprehenderit molestiae vitae similique.

Panel 4 header
Lorem ipsum dolor sit amet, consectetur adipisicing elit. Repellendus aliquid tempore natus voluptates repudiandae, eos dolorem libero inventore, quod incidunt, asperiores. Reiciendis itaque enim pariatur, blanditiis minima autem quo a nulla, obcaecati quis, excepturi atque ab rerum! Sequi molestias vel eum, hic, perspiciatis eius suscipit reprehenderit molestiae vitae similique.
Panel 4 header 2
Lorem ipsum dolor sit amet, consectetur adipisicing elit. Repellendus aliquid tempore natus voluptates repudiandae, eos dolorem libero inventore, quod incidunt, asperiores. Reiciendis itaque enim pariatur, blanditiis minima autem quo a nulla, obcaecati quis, excepturi atque ab rerum! Sequi molestias vel eum, hic, perspiciatis eius suscipit reprehenderit molestiae vitae similique.
Panel 5 header
Lorem ipsum dolor sit amet, consectetur adipisicing elit. Repellendus aliquid tempore natus voluptates repudiandae, eos dolorem libero inventore, quod incidunt, asperiores. Reiciendis itaque enim pariatur, blanditiis minima autem quo a nulla, obcaecati quis, excepturi atque ab rerum! Sequi molestias vel eum, hic, perspiciatis eius suscipit reprehenderit molestiae vitae similique.
Panel 5 header 2
Lorem ipsum dolor sit amet, consectetur adipisicing elit. Repellendus aliquid tempore natus voluptates repudiandae, eos dolorem libero inventore, quod incidunt, asperiores. Reiciendis itaque enim pariatur, blanditiis minima autem quo a nulla, obcaecati quis, excepturi atque ab rerum! Sequi molestias vel eum, hic, perspiciatis eius suscipit reprehenderit molestiae vitae similique.
Panel 5 header 3
Lorem ipsum dolor sit amet, consectetur adipisicing elit. Repellendus aliquid tempore natus voluptates repudiandae, eos dolorem libero inventore, quod incidunt, asperiores. Reiciendis itaque enim pariatur, blanditiis minima autem quo a nulla, obcaecati quis, excepturi atque ab rerum! Sequi molestias vel eum, hic, perspiciatis eius suscipit reprehenderit molestiae vitae similique.